Recently, Yang Chaoyong's team from Institute of Molecular Medicine, Shanghai Jiao Tong University School of Medicine and Zheng Junhua's team from Department of Urology, Renji Hospital published important research results inNature Biotechnology. The study developed Decoder-seq, a highly sensitive and high-resolution spatial transcriptomic sequencing technology, and used it to explore the spatial heterogeneity of the tumor microenvironment of two subtypes of early-stage renal cell carcinoma (RCC), and further resolved the early spatial invasion pattern of tumor cells.
Tumor microenvironment is a highly structured ecosystem with complex components and is closely related to tumor growth, proliferation, and therapeutic response. Therefore, analyzing the tumor microenvironment can help to reveal the composition and spatial distribution of various types of cells in the microenvironment, as well as the complex interactions between tumor cells and other cells, which is of great significance to the study of the mechanisms of tumorigenesis, progression, drug resistance, and so on. In recent years, emerging spatial transcriptome sequencing technologies based on spatial barcode arrays are able to spatially encode tissue sections in situ and provide unbiased, high-throughput whole transcriptome analysis, which provides a powerful tool for tumor microenvironment analysis. However, existing technologies have bottlenecks such as high cost, low sensitivity, and poor resolution.
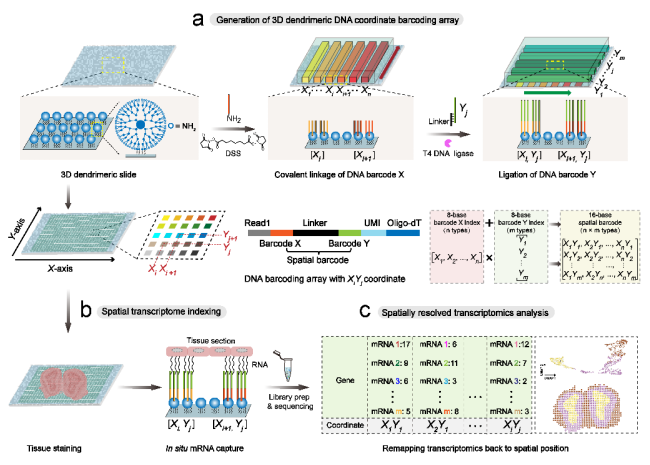
To address these challenges, the research team developed a new spatial transcriptomics technology, Decoder-seq, which utilizes a microfluidic-assisted orthogonal coding strategy to generate arrays of high-density spatial barcodes on 3D nano-substrates, realizing low-cost, highly sensitive, and high-resolution spatial transcriptomics research. First, Decoder-seq increases the modification density of barcodes by about an order of magnitude through 3D tree-like nanosubstrates, which in turn improves the efficiency of tissue mRNA capture and thus increases the sensitivity of gene detection. Second, Decoder-seq utilizes microfluidic-assisted coding to generate coding puncta with flexible spatial resolution (50, 25, 15, and 10 μm). In addition, the deterministic combinatorial barcodes generated by the orthogonal coding strategy significantly reduce the number of coding DNA species and eliminate the need for a decoding step, dramatically reducing experimental costs. Utilizing Decoder-seq with near single-cell resolution (15 μm), highly sensitive detection of ~40 mRNA molecules per μm2 can be achieved, which is much higher than other similar methods. Based on the advantages of Decoder-seq's high sensitivity and high spatial resolution, the spatial distribution resolution of low-expressed olfactory receptor genes in the mouse olfactory bulb and the precise single-cell mapping of the hippocampus were realized.
The research team collected a total of eight tissue samples from the tumor centers and tumor junction areas of three cases of early stage renal clear cell carcinoma (ccRCC) and two cases of early stage renal smoky cell carcinoma (chRCC), and applied Decoder-seq to resolve the tumor microenvironment of the two subtypes of renal cell carcinoma. First, a total of seven cellular ecological niches (CNs) were defined based on cell types and molecular features, revealing significant spatial differences between the two subtypes in terms of cellular compositions, immune infiltration patterns, and cell-cell interactions. Analysis of the immune scores by space revealed that immunosuppression or infiltration was not only associated with the sample subtypes, but also with the spatial distribution of tissues. In addition, it was found that tumor cells in CN5 exhibited a pattern of outward dissemination from the center of the tumor to the invasive margin and then to the normal area, which was consistent with the aggressive growth characteristics of tumor cells. Moreover, there was a strong interaction between tumor cells and immune cells in CN5 in the invasive edge region; whereas in the stroma+normal region showed an up-regulation of depleted state CD8+ T cells, which facilitated tumor cells to evade immune surveillance. Based on the spatial gradient of genes up-regulated in the diffusion direction of CN5, a set of related genes expressing EMT in a spatial gradient was identified (dEMT, 27 genes). dEMT is widely distributed in stromal regions of CN5, suggesting that such regions may serve as "soil" for tumor invasion. Notably, higher dEMT scores were also significantly associated with poorer prognosis and higher tumor stage in RCC, providing important insights into tumor development and treatment.
The study was conducted by a strong collaboration between Chaoyong Yang's team at the Institute of Molecular Medicine, SJTUSM and Junhua Zheng's team at the Department of Urology, Renji Hospital, SJTUSM. Postdoctoral fellow Cao Jiao, PhD student Zheng Chong, and postdoctoral fellow Sun Di of Renji Hospital are the co-first authors; Prof. Yang Chaoyong, Associate Research Fellows Wu Lingling and Song Jia, and Prof. Zheng Junhua are the co-corresponding authors. This study was supported by the National Natural Science Foundation of China, the Key Research and Development Program of the Ministry of Science and Technology of China, and the Innovative Research Team of Shanghai Local High-level University.

