Recently, the research team led by Professor Yu Zhangsheng from the Clinical Research Center of Shanghai Jiao Tong University School of Medicine and the School of Life Sciences and Biotechnology published a paper titled “A foundation model for generalizable cancer diagnosis and survival prediction from histopathological images” inNature Communications. The study presents a foundational pathology model—BEPH (BEiT-based model Pre-training on Histopathological images)—designed for cancer diagnosis and survival prediction, addressing several key limitations in current computational pathology approaches. Master’s student Yang Zhaochang and Assistant Researcher Wei Ting from the School of Life Sciences and Biotechnology are co-first authors; Professor Yu and Associate Researcher Zhang Yue are co-corresponding authors.
In clinical settings, the diagnosis of malignant tumors primarily relies on pathologists examining tissue samples under a microscope. However, manual slide interpretation is time-consuming, heavily dependent on individual expertise, and prone to fatigue and resource limitations—factors that can significantly increase the risk of misdiagnosis or missed diagnoses and delay critical treatment planning. In recent years, computational pathology combined with deep learning has demonstrated tremendous potential in tasks such as cancer detection, subtype classification, and prognosis prediction. These approaches not only significantly enhance diagnostic efficiency but also uncover latent information that traditional methods often miss. Despite this, the development of such technologies faces numerous challenges, including the scarcity of finely annotated data, limited model generalizability, and a lack of interpretability. Moreover, existing models are usually task-specific, requiring retraining or redevelopment for new applications—an issue that adds to the resource burden.
Professor Yu’s team leveraged the Masked Image Modeling (MIM) approach to develop a foundational pathology model—BEPH (BEiT-based model Pre-training on Histopathological images)—specifically designed for cancer analysis and survival prediction. A key advantage of BEPH lies in its ability to perform self-supervised learning on large-scale unlabeled histopathological image datasets, demonstrating strong potential across a wide range of cancer-related tasks. Compared with large, resource-intensive foundation models like CHIEF, BEPH achieves comparable or even superior performance with fewer parameters and limited pretraining data—opening new possibilities for clinical application. (Figure 1)
The model operates in two stages: pretraining and fine-tuning. For pretraining, the team collected approximately11,760 whole-slide images (WSIs) covering 32 cancer types from The Cancer Genome Atlas (TCGA) database, and sampled them into 11.77 million 224×224-pixel image patches. The model was pretrained on this dataset using a self-supervised method MIM. After pretraining, BEPH was tested on various cancer detection tasks to assess its performance and versatility.
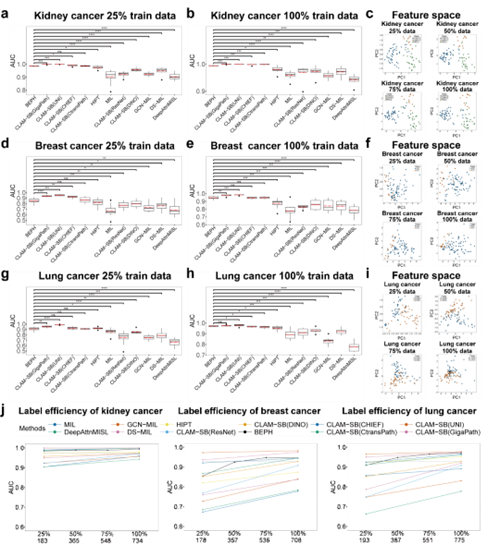
Extensive experiments demonstrated BEPH’s robust adaptability across computational pathology tasks. It significantly improved cancer diagnosis at both the Patch and WSI levels as well as survival risk prediction. For instance, in WSI-level subtype classification tasks across multiple cancer types, BEPH consistently outperformed other weakly supervised models. Even when trained with reduced data, the model retained strong performance, highlighting its practical value in data-scarce clinical environments. (Figure 2)
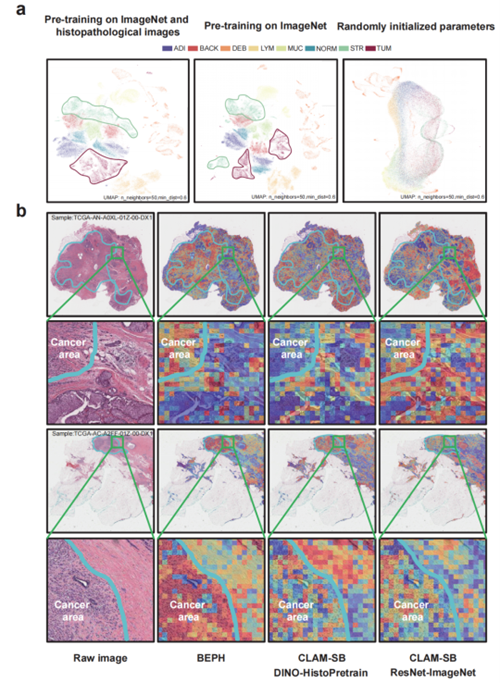
Further heatmap analysis in WSI-level diagnostic tasks showed that BEPH’s attention regions (highlighted in red) aligned closely with cancerous regions annotated by expert pathologists. This indicates that the model can automatically focus on true pathological features. In contrast, other models displayed more scattered attention, with some failing to accurately locate the cancerous regions—suggesting BEPH has stronger capability in identifying relevant pathology. Zoomed-in areas (green boxes) further reveal that BEPH concentrates more precisely on cancerous regions and their boundaries, rather than randomly across the tissue, enhancing the reliability of its decisions. (Figure. 3)
Overall, BEPH offers a unified solution for cancer detection, subtype classification, and survival prediction. Through the dual-phase approach of pretraining and fine-tuning, it efficiently identifies pathological features of cancer, providing powerful support for both diagnosis and prognosis.
This research was supported by the National Natural Science Foundation of China, Science and Technology Commission of Shanghai Municipality, and SJTU’s Interdisciplinary Medical-Engineering Fund, with additional support from the High Performance Computing Center at Shanghai Jiao Tong University.

