Single-Cell Omics in Cancer – A new era for studying tumor biology
Date:2021-11-04 show:
Bulk genomic analyses and expression profiling of clinical specimens have shaped much of our understanding of cancer in patients. However, human tumors are intricate multicellular ecosystems composed of diverse cells, including subsets of cancer, immune, and stromal cells. Their comprehensive characterization cannot be achieved by bulk omics methods.
Single-cell techniques have emerged as powerful approaches to dissect human tumors at single-cell resolution, providing us with a compelling approach to deciphering cancer heterogeneity, biology and crosstalk between the different cellular subsets in the tumor. Emerging studies already highlight the challenges that we are facing for the development of more effective therapeutic strategies. Our cancer program focusses on cancers with unmet medical need, e.g. lung, liver, and pancreatic cancer, and we are addressing the following, non-exclusive questions:
§ Is there a common intra-individual or even inter-individual omics signature?
§ Are current and future therapeutic targets expressed in all clones?
§ What is the spatial distribution of these clones?
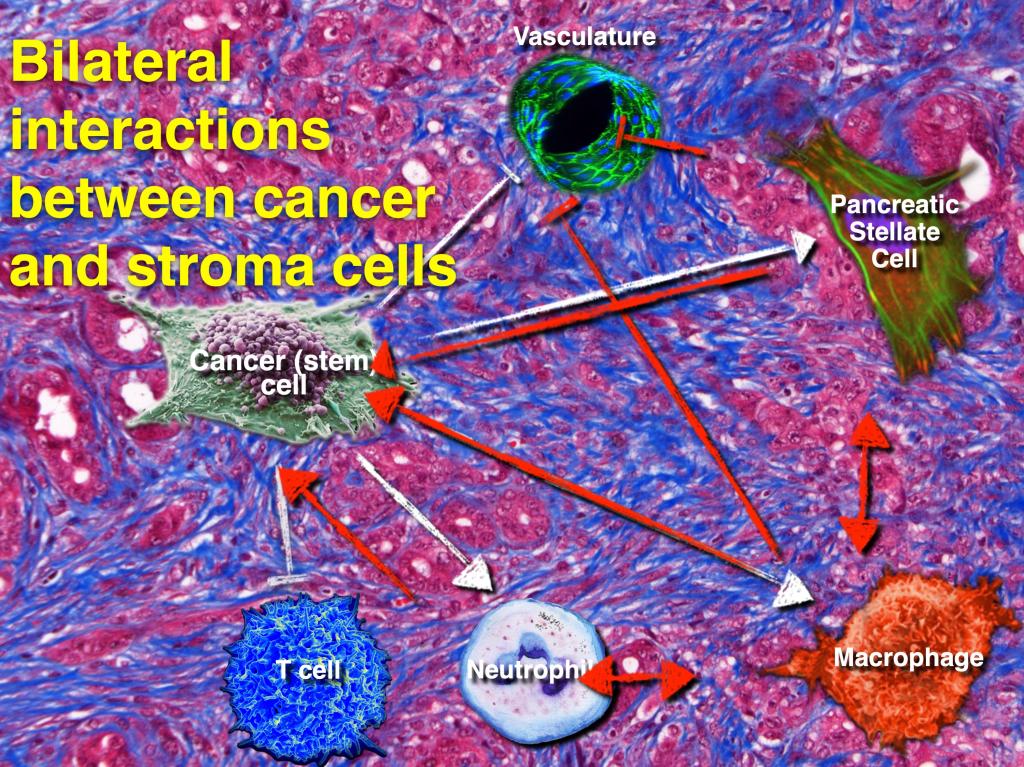
§ What is their (bilateral) relationship with the tumor microenvironment?
§ Can we capture the clonal heterogeneity of cancer cells using liquid biopsies?
§ Are there non-genetic drivers of resistance?
§ Which clones most likely will become dominant and drive relapse of the disease?
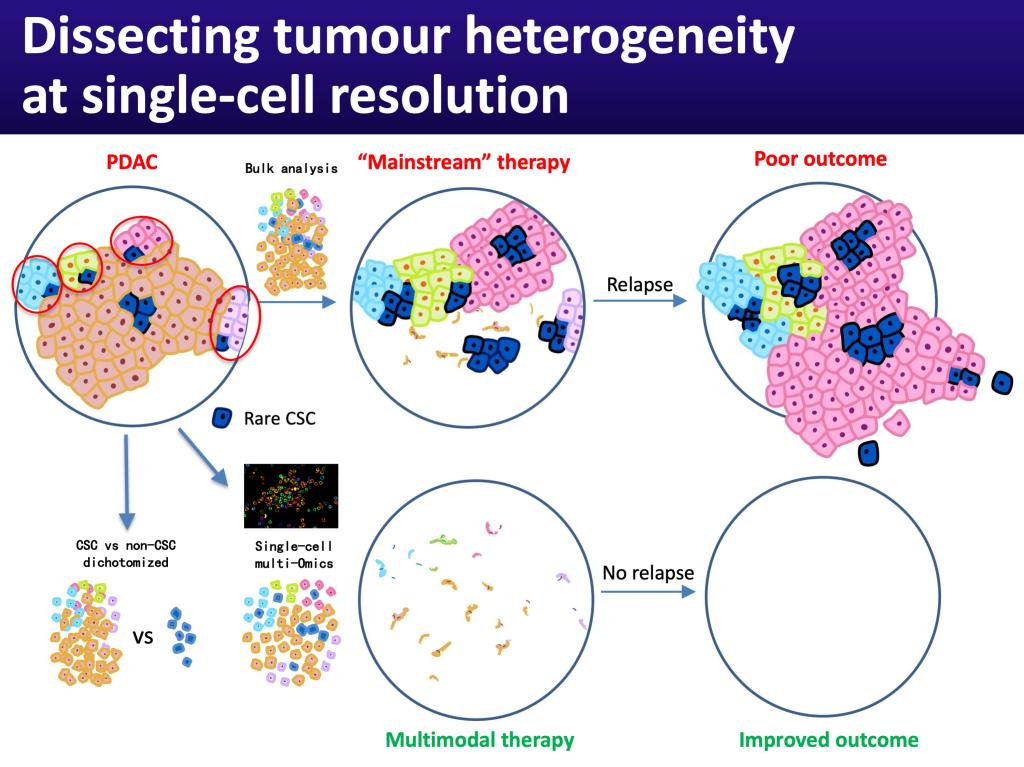
Importantly, cancer heterogeneity is not restricted to clonal heterogeneity, but rather extends to subclonal heterogeneity. Each clone contains a subset of cancer cells that are functionally distinct. These cells are capable of undergoing unlimited cell divisions, while retaining their stem cell identity (self-renewal) and giving rise to highly proliferative progenies, that lack tumor-initiating capacity (differentiation). While these cells do not represent bona fide stem cells as they clearly lack multilineage potential, they do possess certain stem cell traits and therefore have been termed cancer stem cells. These cancer stem cells evolve as the tumor progresses via (epi-)genetic alterations, but also in response to interactions with the tumor microenvironment, leading to diverse cancer stem cell subclones with distinct functionality. In this context, our cancer program aims to answer the following important questions:
§ Can we use single-cell omics technology to detect CSC among non-CSC (and the tumor stroma)?
§ Do CSC also express and depend on current and future therapeutic targets?
§ Is there a common intra-individual or even inter-individual CSC signature that we can explore therapeutically?
§ Can we capture the contained CSC heterogeneity using liquid biopsies?
§ Can we use liquid biopsies as biomarkers for precision medicine approaches?
For more details please check our research programs, e.g. Stem Cells in Cancer Groups

