
郑小琪,博士,教授
上海交通大学公共卫生学院,
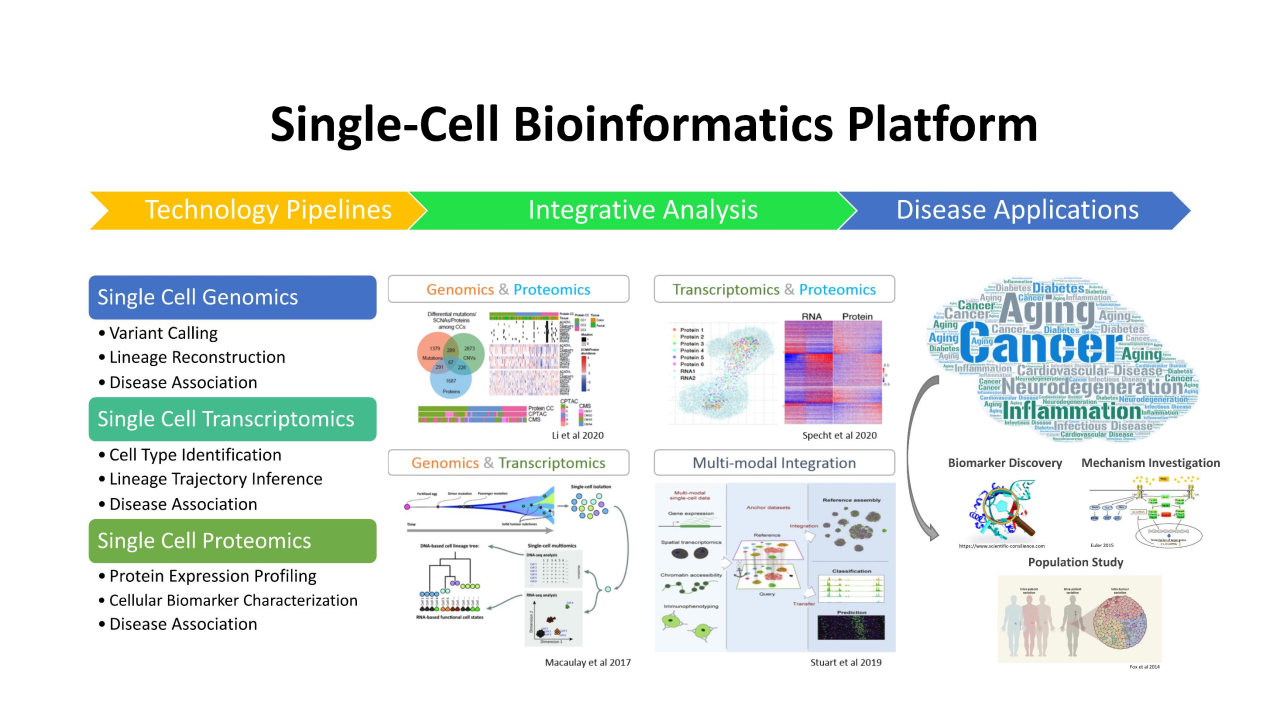
单细胞组学与疾病研究中心,生物信息学平台主任。
通讯地址:上海市黄浦区重庆南路227号
邮箱: xqzheng@shsmu.edu.cn 或 xqzheng@shnu.edu.cn
个人网站: https://xiaoqizheng.github.io/index.html
个人简介:
2009年毕业于大连理工大学应用数学系获理学博士学位,随后在上海师范大学数学系工作,于 2017年晋升至教授、博士生导师。2022年底,加入上海交通大学公共卫生学院,任单细胞组学与疾病研究中心生物信息学平台主任。
主要从事生物统计和生物信息学领域的研究,擅长生物医学大数据的统计建模与算法开发,机器学习、深度学习等人工智能方法在数据挖掘中的应用等。在基于DNA甲基化 (包括亚硫酸盐测序和450k芯片数据) 的肿瘤纯度估计、校正肿瘤纯度的差异甲基化分析及肿瘤样本聚类等问题上取得了一系列研究成果。2008年至今累计发表SCI论文60余篇,包括第一或通讯作者论文40余篇,累积影响因子超过300。主持上海市人才发展基金,以负责人承担国家自然科学基金面上项目2项,青年项目1项,以项目骨干承担国家重点研发计划专项1项。现任中国工业与应用数学学会(CSIAM)数学生命科学分会常务理事,上海非线性科学研究会副秘书长,中国计算机学会生物信息专业委员会委员等。
研究方向:
1. 单细胞转录组与空间转录组数据分析
2. 基于DNA甲基化的肿瘤异质性分析
3. 生物医学研究中的人工智能方法(包括统计学习和深度学习)
教育经历:
2004 - 2009, 博士,应用数学, 大连理工大学
2000 - 2004, 学士,数学与应用数学,山东师范大学
工作经历:
2022.11 – 至今, 教授, 上海交通大学公共卫生学院,单细胞组学与疾病研究中心
2017.09 - 2022.11, 教授, 上海师范大学数学系
2018.07 - 2018.09, 访问学者, 埃默里大学公共卫生学院,生物统计与生物信息学系
2012.09 - 2017.09, 副教授, 上海师范大学数学系.
2012.12 - 2014.02, 访问学者, 哈佛大学公共卫生学院,Dana-Farber癌症研究中心
2009.06 - 2012.09, 讲师, 上海师范大学数学系
代表性论文 (#: 共同第一, *: 共同通讯) :
[1] Wei N, Nie Y, Liu L*, Zheng X*, Wu HJ*, Secuer: Ultrafast, scalable and accurate clustering of single-cell RNA-seq data, PLoS Computational Biology 2022, doi.org/10.1371/journal.pcbi.1010753.
[2] Ding Y, Cai K, Liu L, Zhang Z, Zheng X*, Shi J*, mHapTk: a comprehensive toolkit for the analysis of DNA methylation haplotypes, Bioinformatics. 2022, 38(22):5141-5143.
[3] Zhang Z#, Dan Y#, Xu Y, Zhang J, Zheng X*, Shi J*: The DNA methylation haplotype (mHap) format and mHapTools, Bioinformatics 2021, 37 (24):4892-4894.
[4] Zhang W#, Li Z, Wei N, Wu H-J, Zheng X*: Detection of differentially methylated CpG sites between tumor samples with uneven tumor purities. Bioinformatics 2020, 36(7):2017-2024.
[5] Qin Y#, Zhang W#, Sun X, Nan S, Wei N, Wu HJ, Zheng X*: Deconvolution of heterogeneous tumor samples using partial reference signals. PLoS Computational Biology 2020, 16(11): e1008452.
[6] T Xu, X Zheng, B Li, P Jin, Z Qin, H Wu: A comprehensive review of computational prediction of genome-wide features, Briefings in bioinformatics 2020, 21(1), 120-134.
[7] Liu X, Liu T, Shang Y, Dai P, Zhang W, Lee BJ, Huang M, Yang D, Wu Q, Liu LD, Zheng X, Zhou BO, Dong J, Yeap LS, Hu J, Xiao T, Zha S, Casellas R, Liu XS, Meng FL: ERCC6L2 promotes DNA orientation-specific recombination in mammalian cells, Cell research 2020, 30 (9):732-744.
[8] Hu X, Zhang J, Wang J, Fu J, Li T, Zheng X, Wang B, Gu S, Jiang P, Fan J, Carroll, MC, Wucherpfennig K, Hacohen N, Zhang F, Zhang P, Liu JS*, Li B*, Liu XS*: Widespread B cell clonal expansions and related mechanism of immune evasion in human cancers, Nature Genetics 2019, 51(3):560-567.
[9] Fan H#, Lv P#, Huo X, Wu J, Wang Q, Cheng L, Liu Y, Tang Q, Zhang L, Zhang F, Zheng X, Wu H, Wen B*: The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes, Genome Research, 2018, 28:192-202.
[10] Niu B#, Paulson JN, Zheng X, Kolter R*: Simplified and Representative Bacterial Community of Maize Roots. P Natl Acad Sci USA 2017, 114 (12): E2450-E2459.
[11] Zheng X#*, Zhang N#, Wu HJ, Wu H*: Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biology 2017, 18:17.
[12] Zheng X#*, Zhong S: From structure to function, how bioinformatics help to reveal functions of our genomes. Genome Biology 2017, 18:183.
[13] Zhang W#, Feng H#, Wu H*, Zheng X*, Accounting for Tumor purity improves cancer subtype classification from DNA methylation data, Bioinformatics 33(17), 2017, 2651–2657.
[14] Ni T#, Li XY#, Lu N#, An T, Liu ZP, Fu R, Lv WC, Zhang YW, Xu XJ, Grant Rowe R, Lin YS, Scherer A, Feinberg T, Zheng XQ, Chen BA, Liu XS, Guo QL, Wu ZQ*, Weiss SJ*: Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol 2016, 18:1221-1232.
[15] Wang F#, Zhang N, Wang J, Wu H#, Zheng X*: Tumor purity and differential methylation in cancer epigenomics. Briefings in Functional Genomics 2016, 15:408-419.
[16] Zhang N#, Wu HJ#, Zhang W, Wang J, Wu H*, Zheng X*: Predicting tumor purity from methylation microarray data. Bioinformatics 2015, 31:3401-3405.
[17] Zhang N#, Wang H#, Fang Y, Wang J*, Zheng X*, Liu XS*: Predicting Anticancer Drug Responses Using a Dual-Layer Integrated Cell Line-Drug Network Model. PLoS Computational Biology 2015, 11:e1004498.
[18] Dong Z#, Zhang N#, Li C, Wang H, Fang Y, Wang J, Zheng X*: Anticancer drug sensitivity prediction in cell lines from baseline gene expression through recursive feature selection. BMC Cancer 2015, 15:489.
[19] Zheng X#, Zhao Q#, Wu H-J#, Li W, Wang H, Meyer CA, Qin QA, Xu H, Zang C, Jiang P, Li F, Hou Y, He J, Wang J, Wang J, Zhang P*, Zhang Y*, Liu XS*: MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes. Genome Biology 2014, 15:419.
[20] Li L#, Yu S, Xiao W, Li Y, Huang L, Zheng X*, Zhou S*, Yang H*: Sequence-based identification of recombination spots using pseudo nucleic acid representation and recursive feature extraction by linear kernel SVM. BMC bioinformatics 2014, 15:340.
[21] Zhu J#, Qin Y, Liu T, Wang J, Zheng X*: Prioritization of candidate disease genes by topological similarity between disease and protein diffusion profiles. BMC bioinformatics 2013, 14:S5.
[22] Wang J#, Zheng X*: Comparison of protein secondary structures based on backbone dihedral angles. J Theor Biol 2008, 250:382-387.
单细胞生物信息学平台
平台主任:郑小琪
技术员:胥宛星