Genomics sequencing has become a common approach to study the molecular mechanism and genetic characteristics of diseases. With the continuous progress of technology, single-cell genomics sequencing has become the premium approach to improve the resolution of research. However, single-cell whole genome sequencing generally have the disadvantages of large redundancy of data, low sequencing depth and coverage, difficult data interpretation, and high error rate at present stage, moreover, it is difficult to discriminate true biological variants from technical artifacts, which is not conducive for deep mining of low-abundance variants, and the high cost limits its application in large-scale, high-throughput single-cell studies. Furthermore, though composing a very small fraction (1.1%) of the genome, mutations in the exome are thought to harbor 85% of mutations that have a large effect on disease. Thus, vigorous development of single-cell exome sequencing with higher resolution, higher detection rate, and cost-effectively will substantially improve research efficacy.
Recently, STAR Protocols, a Cell sister journal, onlined the paper entitled "Isolation of single cells from human hepatoblastoma tissues for whole-exome sequencing" contributed by Hui Wang and Jian He’s group.
This strategy demonstrates an ingenious procedure of isolating single cells from tumor tissue and performing single-cell whole exome sequencing. The integration of the single-cell handling and single-cell exome sequencing procedure enabled the improvement of coverage and low-abundance variants detection, also cost-effective, time-saving, and easy to handle. Before that, the team hold several related patents, which mainly focus on single-cell (nuclei) sequencing and single-cell multi-omics analysis.

In this study, He et al. demonstrated a strategy for deciphering the tumor microenvironment of hepatoblastoma using single-cell exome sequencing. An innovative single-cell whole-exome sequencing protocol was developed by integrating single-cell handling with whole-exome sequencing. This work detailed described a series of steps involved in single-cell sorting, whole-genome amplification, amplification uniformity assessment, exome capture, whole-exome library construction, sequencing, and analysis. The uniformity assessment of amplification with genomic multi-loci qPCR could exclude samples with a potential risk of amplification bias before exome capture to guarantee amplification homogeneity and genome coverage. Compared with single-cell whole-genome sequencing, this protocol largely ameliorates the detection rate of low-frequency mutations, enhances the resolution of studies and contributes to genetic heterogeneity study in tumors.
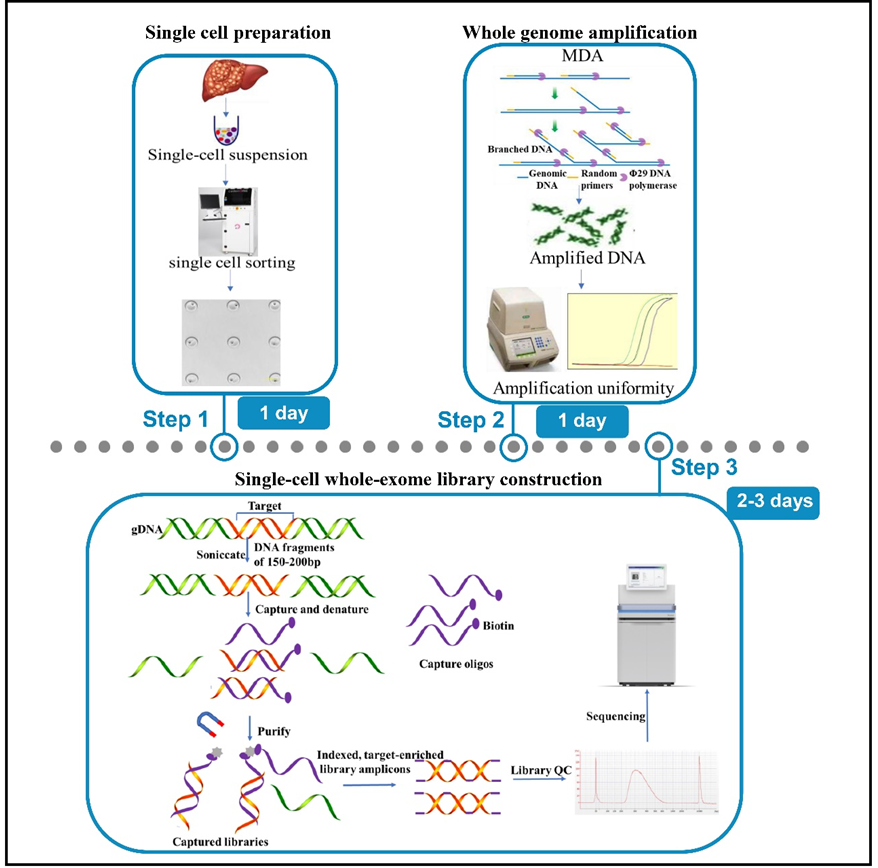
In summary, this paper systematically detailed account of the procedure of single-cell whole exome sequencing using hepatoblastoma samples, and evaluated the effectiveness of this solution through library quality control and sequencing analysis, established the stability and importance of the protocol in detecting low-frequency mutations and revealing the genetic heterogeneity of human diseases. It provides effective reference and guidance for the evolution and ecology of tumor, clinical diagnosis and etc. At the same time, the precautions and trouble-shooting to common troubles during the experimental process and detailed reagents and consumables lists provides in this work enhances the reproducibility, transparency, availability, and repeatability of this protocol.
Professor Hui Wang (Dean of the School of Public Health/ CSCOmics, Shanghai Jiao Tong University School of Medicine) is the corresponding author, and Associate Professor Jian He (Core director of the CSCOmics) and Mei Meng are the co-first authors of this work. This work was supported by the National Key R&D Program of China (2018YFC2000700, 2022YFD2101500, 2022YFD2101503), the National Natural Science Foundation of China (82030099), the Shanghai Science and Technology Commission (22DZ2303000), and the High-level Local University Innovation Research Team.




